About BioTransformer
BioTransformer is an open access software tool, and freely accessible web service for accurate, and comprehensive in silico metabolism prediction and metabolite identification. It makes predicitons using the Metabolism Prediction Tool (BMPT) and the users can then identify the metabolites of their interest using the filters provided on the result page. The current version of the BioTransformer Rails application is 3.1.0.
Structure and Implementation
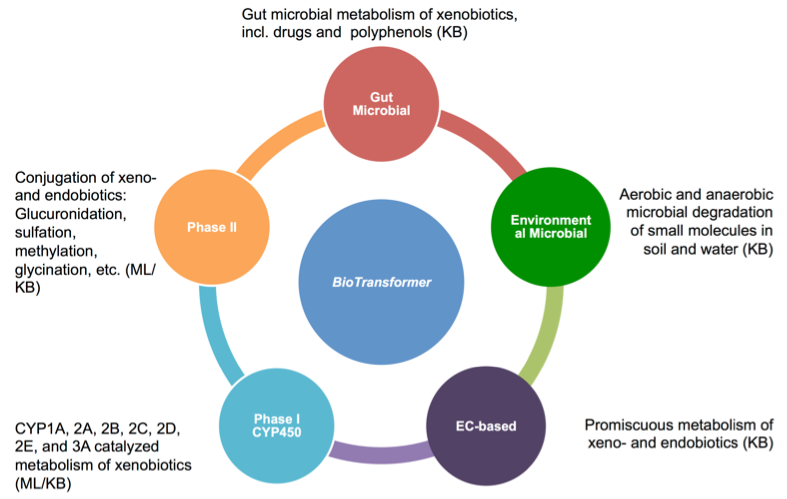
The BMPT consists of five independent prediction modules called “transformers”, namely:
- the EC-based transformer
- the CYP450 (phase I) transformer
- the phase II transformer
- the human gut microbial transformer
- the environmental microbial transformer (See Fig. 1).
- the abiotic transformer
- a biotransformation database (called MetXBioDB) containing detailed annotations of experimentally confirmed metabolic reactions
- a reaction knowledgebase containing generic biotransformation rules, preference rules, and other constraints for metabolism prediction
- a reasoning engine that implements both generic and transformer-specific algorithms for metabolite prediction and selection.
Evaluation
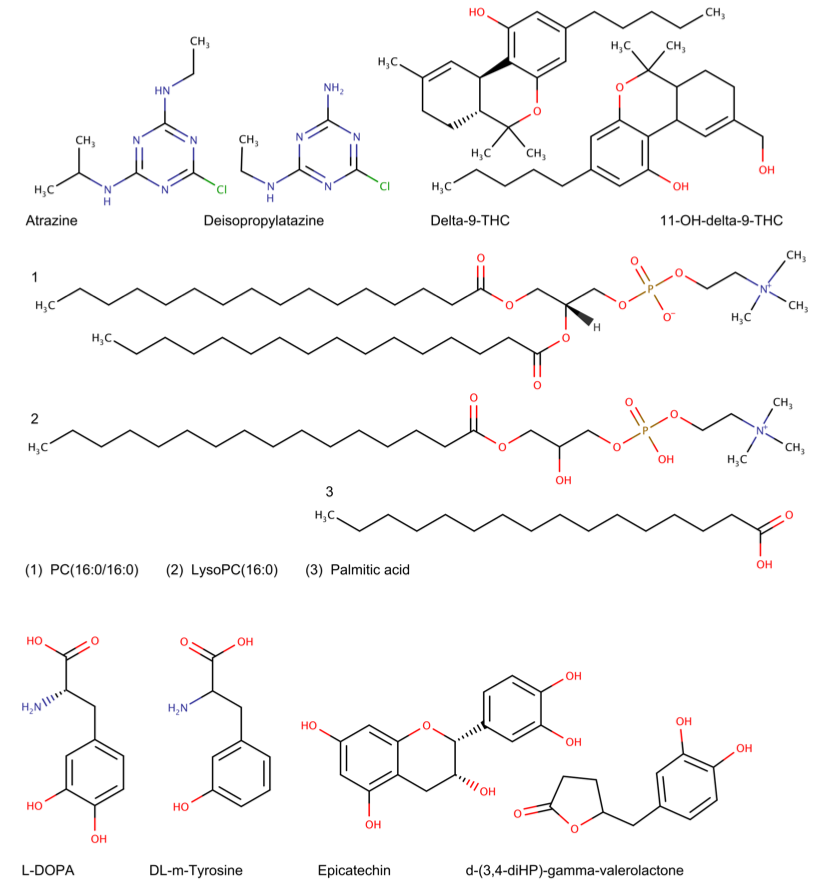
A comprehensive evaluation of BioTransformer showed that it was able accurately predict human and human gut metabolism of a diverse set of compounds (See Fig. 2), ranging from endogenous molecules (e.g. glycerophospholipids) to xenobiotics (e.g. pharmaceuticals, pesticides, and other environmental contaminants). Moreover, it was able to outperform two state-of-the-art commercially available tools (Meteor Nexus and ADMET Predictor), with precision and recall values up to 7 times better than those obtained for Meteor Nexus or ADMET Predictor on the same sets of pharmaceuticals, pesticides, phytochemicals or endobiotics under similar or identical constraints. Furthermore BioTransformer was able to reproduce 100% of the transformations and metabolites predicted by the EAWAG Pathway Prediction system.
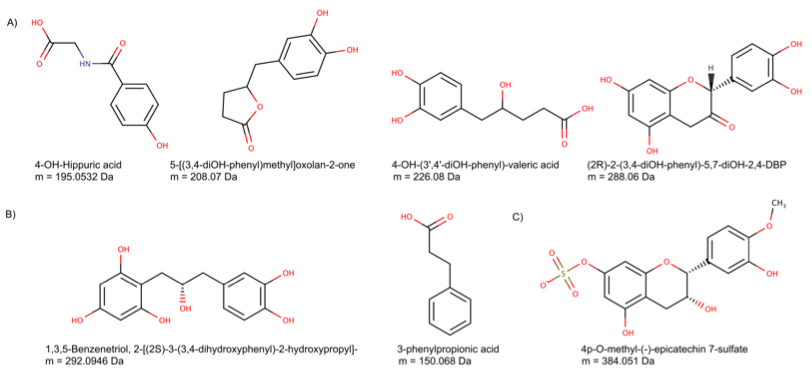
Using mass spectrometry data obtained from a rat experimental study with epicatechin supplementation, BioTransformer was also able to correctly identify 39 previously reported epicatechin metabolites via its Metabolism Identification Tool, and suggest 28 potential metabolites, 17 of which matched nine monoisotopic masses for which no evidence of a previous report could be found (See Fig. 3).